

Wij zijn geïnteresseerd in kleine RNA’s, zogenaamde microRNA’s of miRNA’s, welke helpen om de productie van eiwitten in cellen te reguleren. Ons doel is om hun bijdrage tot een aantal aspecten van metabole ziekten en kanker beter te begrijpen.
Regulatie van eiwitproductie - miRNAs en transcriptiefactoren
Vrijwel alle cellen in het lichaam zijn dragers van dezelfde genetische informatie in de chemische vorm van DNA ( hierna "genen” genoemd) . In een bepaald celtype is slechts een zeer kleine fractie van alle genen “in gebruik". De specifieke selectie van die genen die worden gebruikt in de verschillende celtypen bepaalt in hoge mate hoe een cel eruit ziet en hoe zij zich gedraagt. Dit verklaart waarom bijvoorbeeld een huidcel er heel anders uitziet en zich heel anders gedraagt dan een bloedcel.
Vele stappen zijn er nodig in het proces dat leidt van de genetische informatie (zoals opgeslagen in de vorm van DNA in de celkern) tot aan de productie van eiwitten, die dienen als de cellulaire bouwstenen en machines. Eerst wordt de genetische informatie in de kern gekopieerd naar een kortstondig tussenproduct, het zogenaamde messenger RNA (mRNA). Het mRNA wordt vervolgens van uit de kern naar het cytoplasma geëxporteerd, waar het dient als een matrijs voor de productie van eiwitten.
De regulering van de eiwitproductie vindt plaats op verschillende niveaus. Wij zijn geïnteresseerd in twee niveaus van deze regeling. Ten eerste zijn we geïnteresseerd in hoe een groep van eiwitten, transcriptiefactoren genaamd (met inbegrip van de tumor suppressor factor p53), de productie van mRNA reguleert. Ten tweede, zijn we vooral geïnteresseerd in een groep van regulerende RNA’s, microRNA’s of miRNA’s genaamd, die mRNA's destabiliseren en zo de productie van bepaalde eiwitten verminderen. Chemisch zijn deze miRNA’s opgebouwd uit dezelfde bouwstenen als mRNA, maar ze zijn veel korter zoals hun naam, “micro” RNA (vaak afgekort tot miRNA), aangeeft.
Het menselijk genoom bevat ongeveer duizend miRNA’s. Vreemd genoeg regelt ieder afzonderlijk miRNA niet alleen de productie van één, maar van vaak honderden proteïnen tegelijk. Op hun beurt kunnen veel eiwitten worden gereguleerd door verschillende miRNA’s. Om een inzicht te verkrijgen in de functies van specifieke miRNA’s is dus geen eenvoudige zaak.
In ons werk proberen we te begrijpen hoe deze regulerende processen samen werken in processen als kanker en stofwisseling. Uiteindelijk kan dit ons helpen om nieuwe therapeutische toepassingen te vinden.
Introduction
miRNAs are small regulatory RNAs
miRNAs are small non-protein-coding RNAs that can bind to mRNA transcripts of protein coding genes. Upon binding to these mRNAs, they inhibit their translation into proteins. However, each miRNA does not only recognize one target transcript, but rather numerous – in some cases several hundreds – of target transcripts. In addition, for many miRNAs, multiple different genes exist, that encode highly similar or identical mature miRNAs. The potential for combinatorial complexity and functional redundancy is therefore enormous.
The focus of our group is on the role of miRNAs in conserved signaling pathway at the crossroad of cancer and metabolism.
Specifically, we are pursuing the following lines of investigation:
Regulation of cholesterol and fatty acid metabolism by the bifunctional locus SREBF2-miR33
miRNAs in intestinal differentiation and tumorigenesis
Metabolic changes downstream of p53
miRNAs regulate stability and translation of mRNAs
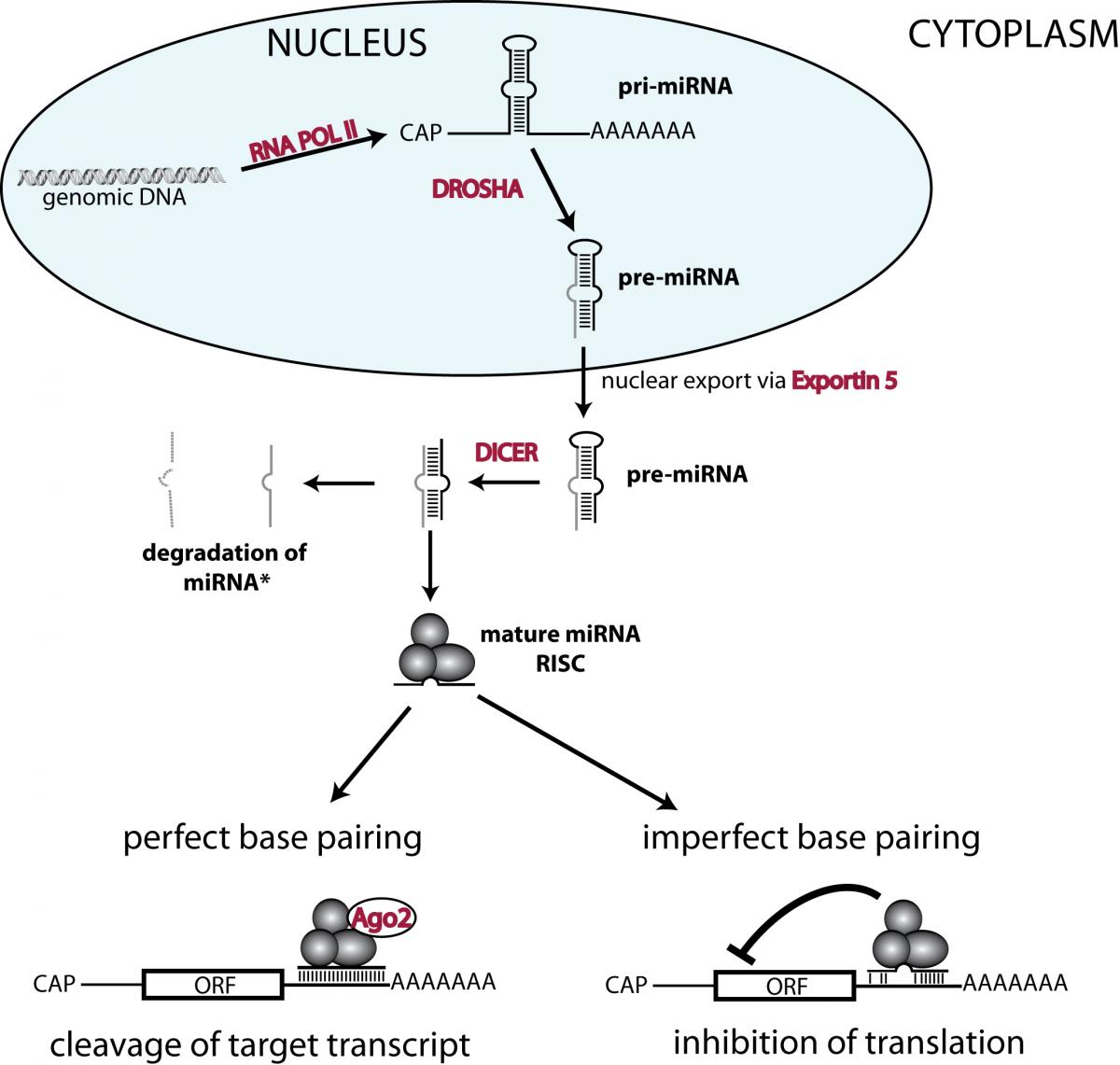
Legend:
Precursor transcripts of miRNAs (i.e. pri-miRNAs) are produced by RNA polymerase II in the nucleus. These transcripts contain hairpin structures that are cleaved out by a complex containing the endonuclease DROSHA. The resulting pre-miRNA is exported from the nucleus into the cytoplasm, where the endonuclease DICER cleaves off the loop and generates double stranded RNAs. One (or sometimes both) of these strands get incorporated in the RNA induced silencing complex (abbreviated as RISC). Depending on the degree of similarity, miRNA binding to the 3’ untranslated region (of target transcripts leads to either AGO2 dependent cleavage or translational inhibition.
ORF= open reading frame; CAP = 5’ mRNA CAP.
Regulation of cholesterol and fatty acid metabolism by the bifunctional locus SREBF2-miR33 (Laure-Alix Clerbau & Benoit Ury, in collaboration with Jennifer Kennell, Vassar College)
Fatty acids, cholesterol, and their lipid derivatives play essential roles in normal cellular function and serve as structural components, signaling molecules, and/or as storage forms of energy. In multicellular organisms, cellular lipid metabolism is regulated to match the needs both of individual cells and of the entire organism.
The sterol regulatory element-binding factor-2 (SREBF2) gene is a bifunctional locus encoding SREBP-2, a well-known transcriptional regulator of genes involved in cholesterol and fatty acid biosynthesis, and miR-33a. We and others have recently shown that miR-33a can reduce the expression of several proteins involved in the cellular export of cholesterol and β-oxidation of fatty acids, thus adding an unexpected layer of complexity and fine-tuning to regulation of lipid homeostasis. In fact, work of other groups has demonstrated that this mechanism might represent a therapeutic target in the treatment of hypercholesterolemia. We are continuing to investigate the physiological role of miR-33 family members in different experimental systems throughout evolution.
The bifunctional locus of SREBF2-miR33 regulates cholesterol and fatty acid metabolism
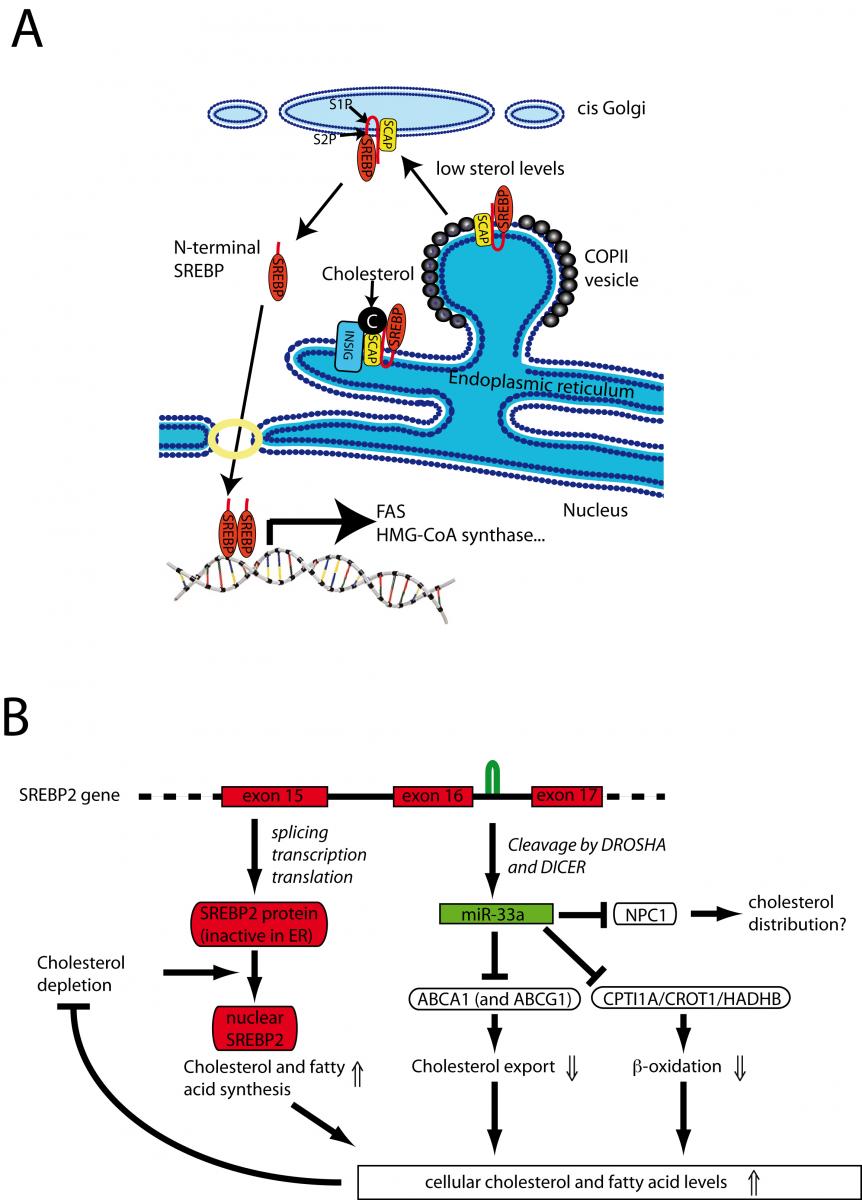
Legend:
After processing from an intron of SREBF2, miR-33a reduces cellular cholesterol export by inhibiting expression of ABCA1 (and in the mouse ABCG1). In addition, miR-33a reduces mitochondrial fatty acid b-oxidation via inhibition of HADHB, CROT, and CPT1A to increase intracellular lipid levels. Thus the SREBF2 locus uses two distinct mechanisms to maintain lipid homeostasis: regulated transcriptional activity of SREBP-2 and translational repression by miR-33a.
miRNAs in intestinal differentiation and tumorigenesis (Jennifer Bolsee, Mathias Halbout, Isabelle Gerin)
The intestine is required for the digestion and absorption of essential nutrients and water. In this process, its surface epithelium is exposed to one of the most toxic milieus of the whole body. It has to resist aggressive digestive juices, large pH changes, anaerobic bacteria and numerous toxic compounds. To resist this, its surface epithelium is completely renewed in less than 2 weeks. An intricate network of signaling pathways controls the proliferation and differentiation from intestinal stem cells to the mature cell types. We are studying the role of miRNAs in this differentiation process and how they contribute to intestinal differentiation as well as the development of colorectal cancer.
Intestinal architecture is maintained by the interplay of many signaling pathways.
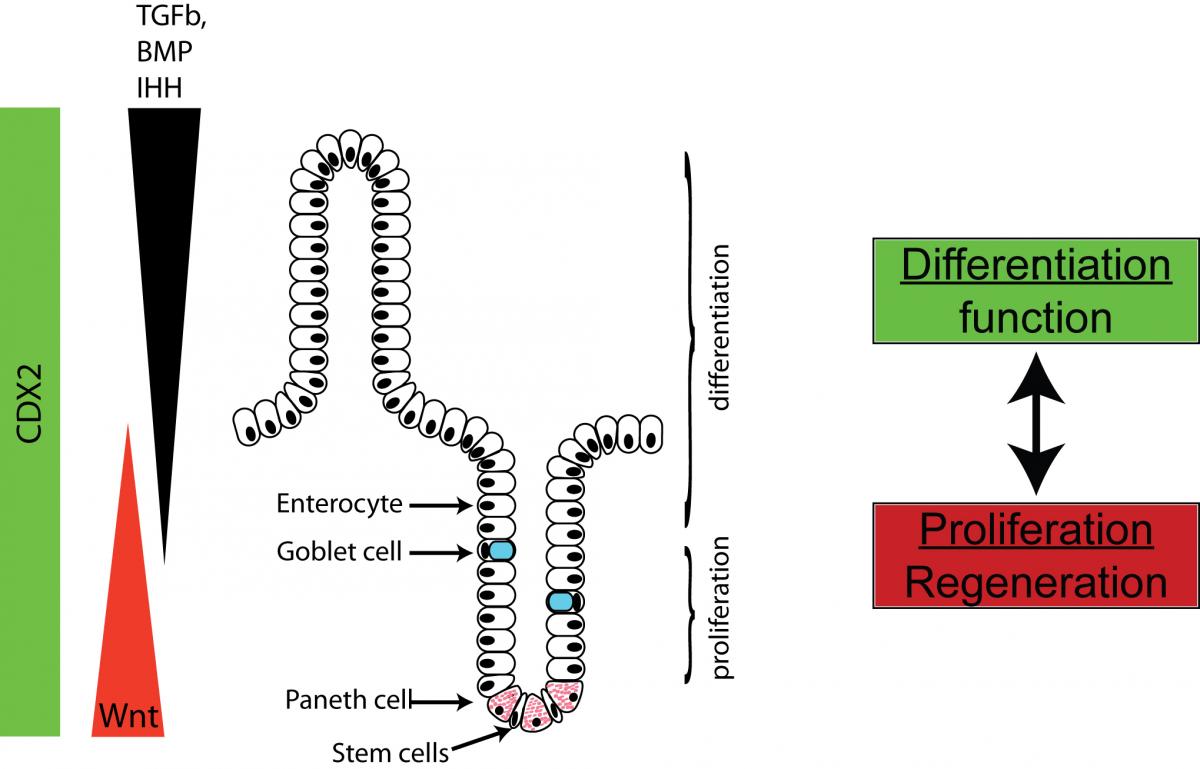
Legend:
The intestinal architecture is maintained by the interplay of signaling patways that ensure complete renewal of intestinal surface epithelia within 7 to 14 days. New cells are generated from a stem cell compartment at the base of the crypts and successively migrate up, where they are eventually shed in the lumen. We are focusing our interest on miRNAs that regulate this process.
Metabolic changes downstream of p53 (Isabelle Gerin, Jennifer Bolsee)
The p53 protein is a transcription factor that acts as the central hub of a tumor suppressor pathway. It is stabilized when cells are exposed to a large number of stresses such as oxidative stress, nutrient starvation, oncogene activation and DNA damage. This leads to the transcriptional activation of many target genes. Upon mild stress, these target genes induce cell cycle arrest, reduce oxidative stress and activate DNA damage repair pathways. This seems to help sub-lethally damaged cells to repair the incurred insults. Upon stronger stress, cell death and permanent cell cycle arrest (or senescence) remove damaged cells from the proliferative pool. In recent years, the effects of p53 on metabolism and cellular redox status have received increasing attention. One of the milestones was the identification of the gene TIGAR as direct target gene of p53. As indicated by its name Tp53 induced glycolysis and apoptotis regulator), this protein regulates glycolysis and modifies apoptosis. It has long been assumed that this effect is due to a direct effect of TIGAR on the glycolytic regulator fructose 2,6-bisphosphate. We have recently found that fructose 2,6-bisphosphate is a rather poor substrate for TIGAR (kcat/Km 0.3 mM-1 s-1) and that 2,3-bisphosphoglycerate is an about 400x better substrate (kcat/Km 135 mM-1 s-1).
This has significant implications for our understanding of the metabolic changes downstream of p53 as well as for cancer cell metabolism in general. It also suggests that the role of 2,3-bisphosphoglycerate needs to be revisited.
TIGAR and metabolic adaptations in cancer
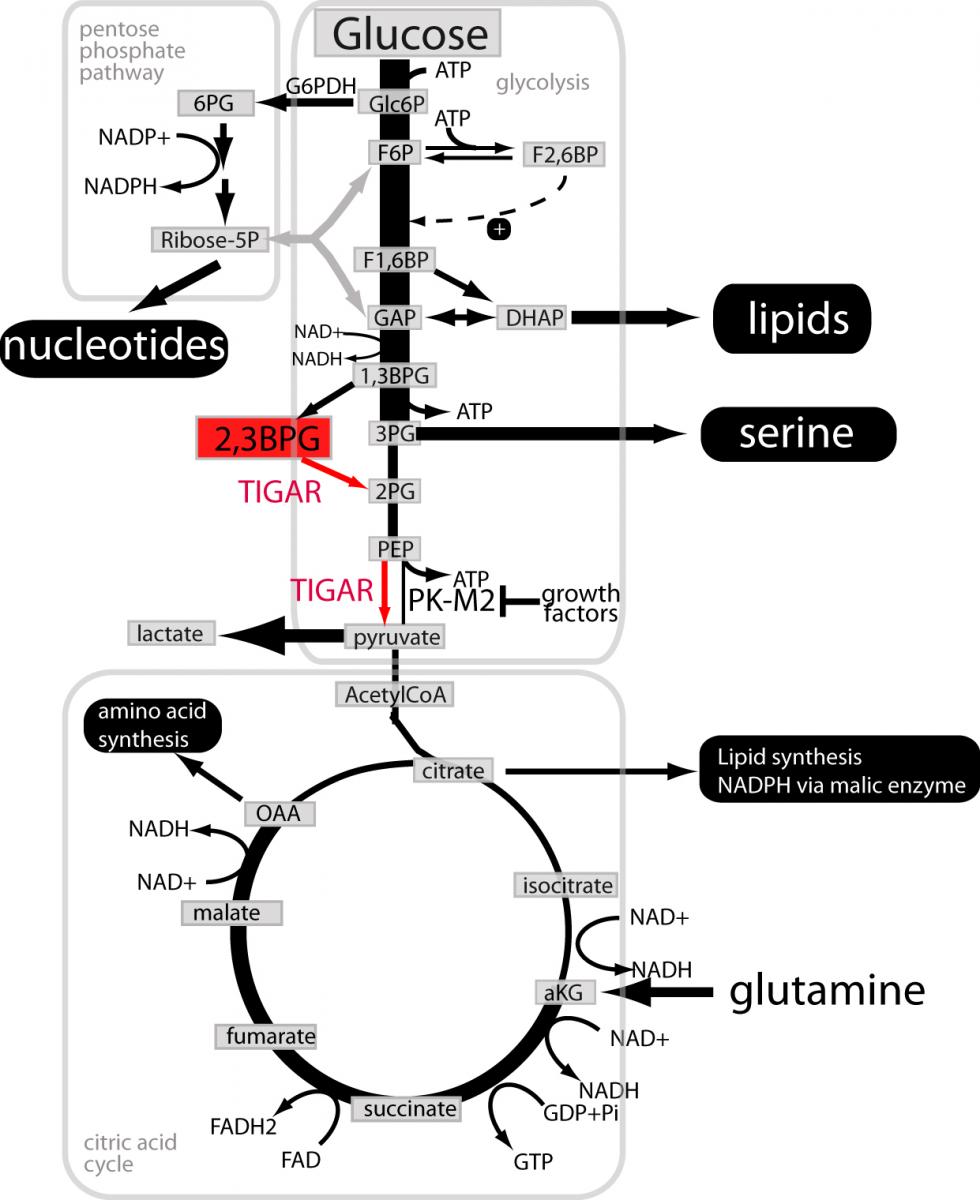
Legend:
The local “success” of a cancer cell is measured by its ability to proliferate and survive better with the available nutrients than its neighbouring normal or cancerous cells. Like any other cell, a cancer cell needs to maintain cellular integrity and fulfill baseline housekeeping functions (reviewed in Vander Heiden et al 2011). Many thermodynamically unfavourable reactions are driven by concomitant hydrolysis of ATP. All cell types therefore need to synthesize ATP by breaking down nutrients in pathways such as glycolysis, citric acid cycle and most efficiently mitochondrial oxidative phosphorylation. In addition, proliferating cells in general and cancer cells in particular need to generate bio-mass. Synthesis of amino acids, nucleotides and lipids starts with precursors that are intermediary products in the same pathways that are used to synthesize cellular ATP. Several adjustments of the flux through these pathways are needed to reconcile cellular demand for biosynthetic building blocks and for ATP synthesis. Our data suggest that TIGAR might contribute to the metabolic adaptations in cancer at the indicated steps.
Nat Chem Biol. 2016; 12(8):601-7.
Nat Commun. 2016; 7:11534.
Gerin I, Noël G, Bolsée J, Haumont O, Van Schaftingen E, Bommer GT.
Biochem J. 2014; 458(3):439-48.
Feng Y, Bommer GT, Zhao J, Green M, Sands E, Zhai Y, Brown K, Burberry A, Cho KR, Fearon ER.
Gastroenterology. 2011; 141(3):1003-1013.e1-10.
Bommer GT, MacDougald OA.
Cell Metab. 2011; 13(3):241-7.
Fearon ER, Bommer GT.
Gastroenterology. 2011; 140(4):1139-43.
Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT.
J Biol Chem. 2010; 285(44):33652-61.
Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA.
Am J Physiol Endocrinol Metab. 2010; 299(2):E198-206.
Herbst A, Bommer GT, Kriegl L, Jung A, Behrens A, Csanadi E, Gerhard M, Bolz C, Riesenberg R, Zimmermann W, Dietmaier W, Wolf I, Brabletz T, Göke B, Kolligs FT.
Gastroenterology. 2009; 137(2):639-48, 648.e1-9.
Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER.
Curr Biol. 2007; 17(15):1298-307.

p53 EN miRNA’s IN KANKER EN METABOLISME