

Our objective is to understand why the immune response that cancer patients mount against their tumour fails to control its progression, and how we can revert this. To this aim, we study the interaction between immune cells and cancer cells in human tumor samples.
Cancer cells carry antigens on their surface that allow the immune system to distinguish them from normal cells and to destroy them. Many cancer patients mount immune responses against their cancer, as evidenced by the inflammatory reactions that are frequently observed in tumors. These spontaneous responses, even though they may help slow down and maybe even in some cases eradicate some tumors, are clearly ineffective against metastatic disease. It is likely that advanced tumors select, under pressure from the immune system, mechanisms that enable them to resist killing. It is important to identify these resistance mechanisms so as to develop countermeasures capable of restoring effective anti-tumor immunity.

Our laboratory studies the active immune responses that occurs in tumors. We focus on melanoma, colorectal tumors and lymphomas. These tumors are frequently infiltrated by immune cells such as lymphocytes and macrophages. We use several complementary techniques, combining microscopy and genetic analysis, to study the details of these responses, such as the activation status of these cells, and thus attempt to highlight immune dysfunctions that could shed light on the mechanisms of resistance that are active in these tumors.
In practice, operated or biopsied tumor fragments are frozen, then cut into very thin slices. They undergo several stainings that highlight the cells and allow to distinguish tumor from immune cells. We can then determine the number and distribution of immune cells, as well as certain of their functional characteristics (e.g. if they have an activated status). It is possible to see if the tumor cells are proliferating, or if they are dying. The study of expressed genes allows to determine the type of inflammation that is active in tumors. By combining these analyzes, we can specify whether the immune cells are activated in the tumor, or rather if they are inhibited by the tumor environment.
Human tumors express a wide diversity of tumor antigens that are not or poorly expressed on normal tissues and that can be recognized by cytolytic T lymphocytes (CTL). Cancer patients mount spontaneous CTL responses against their tumor, as indicated by the presence of anti-tumor CTL circulating in their blood or infiltrating their tumor, sometimes in high numbers. However, these responses are not capable to control tumor growth in patients with advanced disease. They can fail at different steps: absence of tumor antigens or appropriate antigen presentation, absence of CTL activation or amplification, failure for activated CTL to migrate to and infiltrate the tumor tissue, impaired T cell killing. By studying the pattern, abundance and phenotype of tumor-infiltrating CTL in situ, in human tumor samples, we try to understand this failure.
Characterization of T and B-cell responses in melanoma metastases
In collaboration with the group of P. Coulie (Cellular Genetics, de Duve Institute)
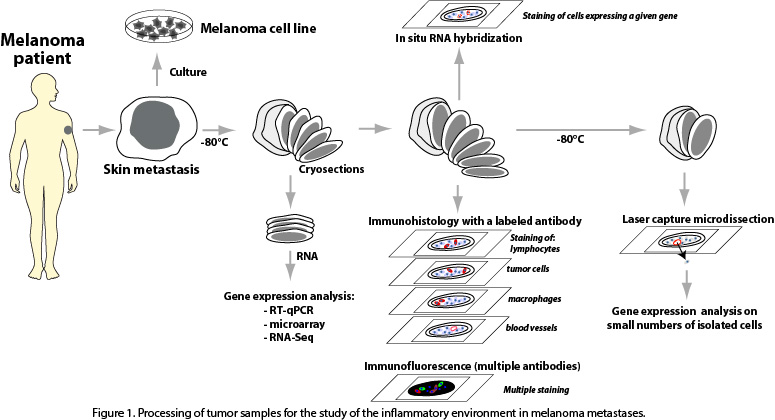
We study the immune and inflammatory components and their interaction with tumor cells in freshly resected cutaneous metastases obtained from melanoma patients. Part of the resected tumors are put in culture, in order to attempt to derive immortalized melanoma cell lines, which are precious tools in experimental tumor immunology. The remaining piece of tumor is kept frozen. Thin tissue sections are cut from this material, and are used for RNA extraction followed by gene expression profiling, for immunohistochemistry and immunofluorescence stainings, for in situ RNA hybridization and for laser capture microdissection of small tissular regions of interest (Fig. 1).
These complementary approaches are aimed at allowing us to characterize the mechanisms by which the tumors resist to their destruction by the anti-tumoral immune responses that they trigger.
We focus on the following elements to investigate intra-tumoral T cell responses:
- We study functional gene expression signatures observed in microarray and whole transcriptome profiling. We have identified a signature that comprises T cell activation genes, CTL genes and IFN-γ target genes, and reflects Th1-oriented T cell activation. We also assess the expression of phenotypic markers of T cell activation in situ.
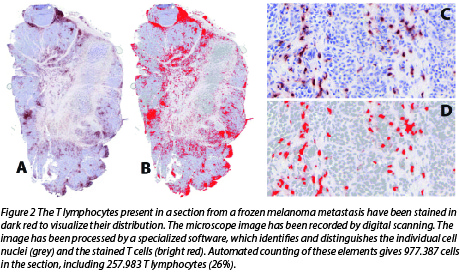
- We study the abundance and pattern of T cell distribution in the tumors, and their spatial relationship with the tumor cells. This is illustrated in Figure 2.
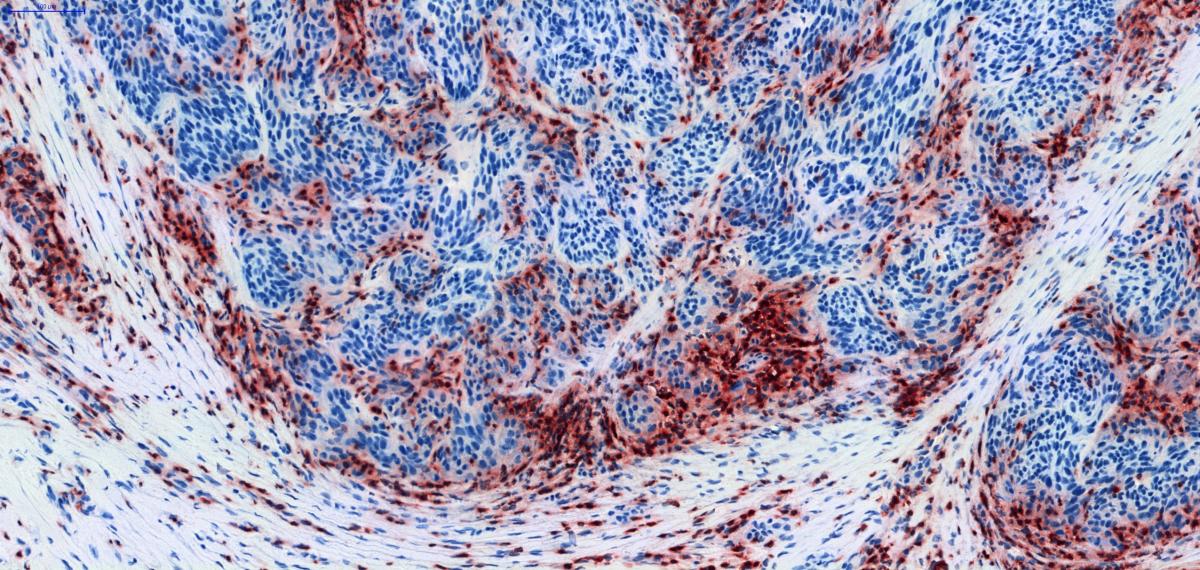
- We study markers of tumor cell killing by cytolytic T cells, such as molecules that become expressed following activation of the T cell lytic machinery or are associated with apoptosis of tumor cells. The aim is to assess whether tumor cell killing occurs, in which tumors, and where precisely.
- As some tumors are known to resist immune rejection by losing or down-modulating proteins required for antigen processing and presentation, we study the expression of the main components of the antigen presentation machinery (APM) in our tumor samples. An example is shown in Fig.3.
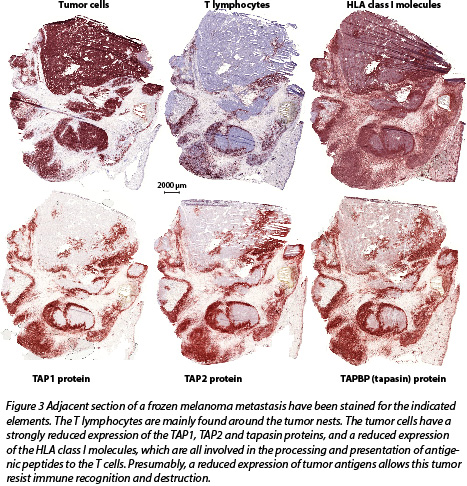
- We assess whether these different markers are correlated with each other and with the clinical outcome of the patients.
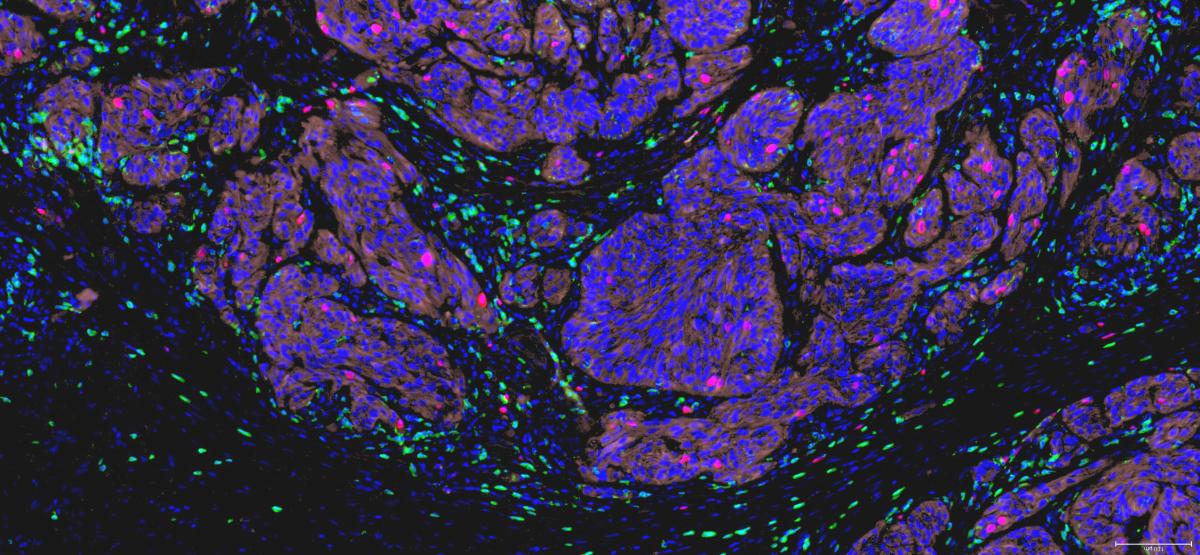
We have observed the presence of ectopic lymphoid structures, also called tertiary lymphoid organs, in melanoma metastases. These structures are organized in B-cell follicles, adjacent T cell areas and neighbouring high endothelial venules, and thus contain the main components required to support local adaptive B and T cell responses. The presence of germinal centers and the occurrence of immunoglobulin affinity maturation in some follicles reveals ongoing B cell responses. The intimate association of mature dendritic cells and T lymphocytes in the T cell areas suggests that T cell responses also take place inside these structures (Fig. 4). This phenomenon, called lymphoid neogenesis, is frequently observed in various chronic inflammatory diseases. It has also been described in several types of tumors, including breast, lung and testis cancer. It is a consequence of sustained lymphocyte activation in the presence of persistent antigenic stimuli.
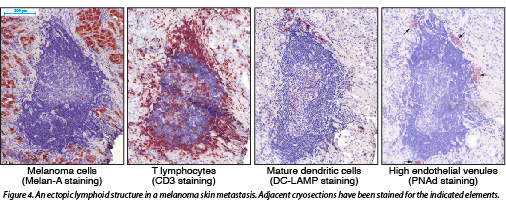
Altogether, our observations suggest that the melanoma environment is the site of sustained immune activity rather than of a widespread immune suppression as is frequently proposed.
Characterization of tumor antigens and T-cell responses in diffuse large B-cell lymphoma
In collaboration with A. Camboni (Department of Pathology), E. Van Den Neste (Hematology), C. De Smet and A. Loriot (de Duve Institute)
Diffuse large B-cell lymphoma (DLBCL) can be distinguished from the other tumors by its propensity to occur in severely immunosuppressed individuals and by its frequent genetic loss of antigen-presenting molecules, indicating that it is a privileged target for T-cell responses. The reason for this is unclear. The antigenic targets of these responses are unknown.
Using the same approaches as those developed in melanoma, we study large series of DLBCL samples. We focus on the loss of expression of HLA class I heavy chain molecules and beta-2-microglobulin, which are particularly frequently observed in this malignancy. We also study tumoral expression of PD-L1, which binds and inhibits PD-1, a T-cell co-stimulatory molecule, and loss of CD58/LFA-3, the ligand of the CD2 T-cell and NK cell co-stimulatory molecule, both of which have been reported in DLBCL. These results are correlated with the CD4 and CD8 T-cell infiltration pattern in the tumor, and with clinical and biological parameters.
Study of IDO1 expression in human malignancies
In collaboration with the group of B. Van den Eynde, A. Mourin (Department of Pathology), M. Van den Eynde (Medical Oncology) and the Biobank (Cliniques universitaires Saint-Luc, Brussels)
Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 (IDO1) plays an important role in tumor resistance to immune rejection (see the report of B. Van den Eynde). In humans, constitutive expression of IDO1 has been observed in several tumor types. The precise expression profile of IDO1 in human tissues has remained unclear, due to the use of different methodologies and antibodies used in immunostaining assays, including antibodies with doubtful specificity. A precise profiling is important to assess the risks and potential benefits of IDO1 inhibitors, which are currently in pre-clinical and early clinical development, and to identify the best tumor targets for this treatment.
We have performed an extensive immunohistochemical analysis of IDO1 expression in normal and tumor tissues. In normal tissues, IDO1 was expressed by endothelial cells in placenta and lung, and by epithelial cells in the female genital tract. It was also detected in lymphoid tissues, in interstitial cells corresponding to mature dendritic cells. IDO1-expressing cells were observed in a large fraction (505/866, 58%) of human tumors. Tumors showing the highest proportions of IDO1-immunolabelled samples included carcinomas of endometrium and cervix, followed by kidney, lung, and colon. Other tumors such as glioblastomas were often negative. This hierarchy was confirmed by gene expression data mined from the TCGA database. IDO1-positive cells were tumor cells, endothelial cells and stromal cells, in proportions that varied depending on the tumor type (Fig. 5).
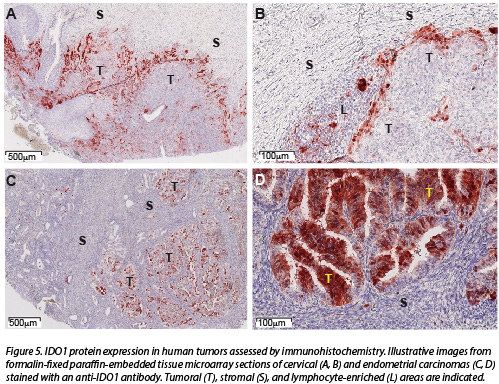
Based on these results, we now focus on colorectal cancer, and analyze a larger series of tumor samples in greater detail. Next to IDO1 expression, we analyze its spatial relationship with T cell distribution in tumors and with the clinical evolution of the patients.
Nat Commun. 2017; 8(1):1404.
Oncoimmunology. 2015; 4(5):e1003012.
Loriot A, Van Tongelen A, Blanco J, Klaessens S, Cannuyer J, van Baren N, Decottignies A, De Smet C.
Epigenetics. 2014; 9(8):1163-71.

IMMUNE AND INFLAMMATORY ENVIRONMENT IN HUMAN TUMORS